
What is DeepCas13
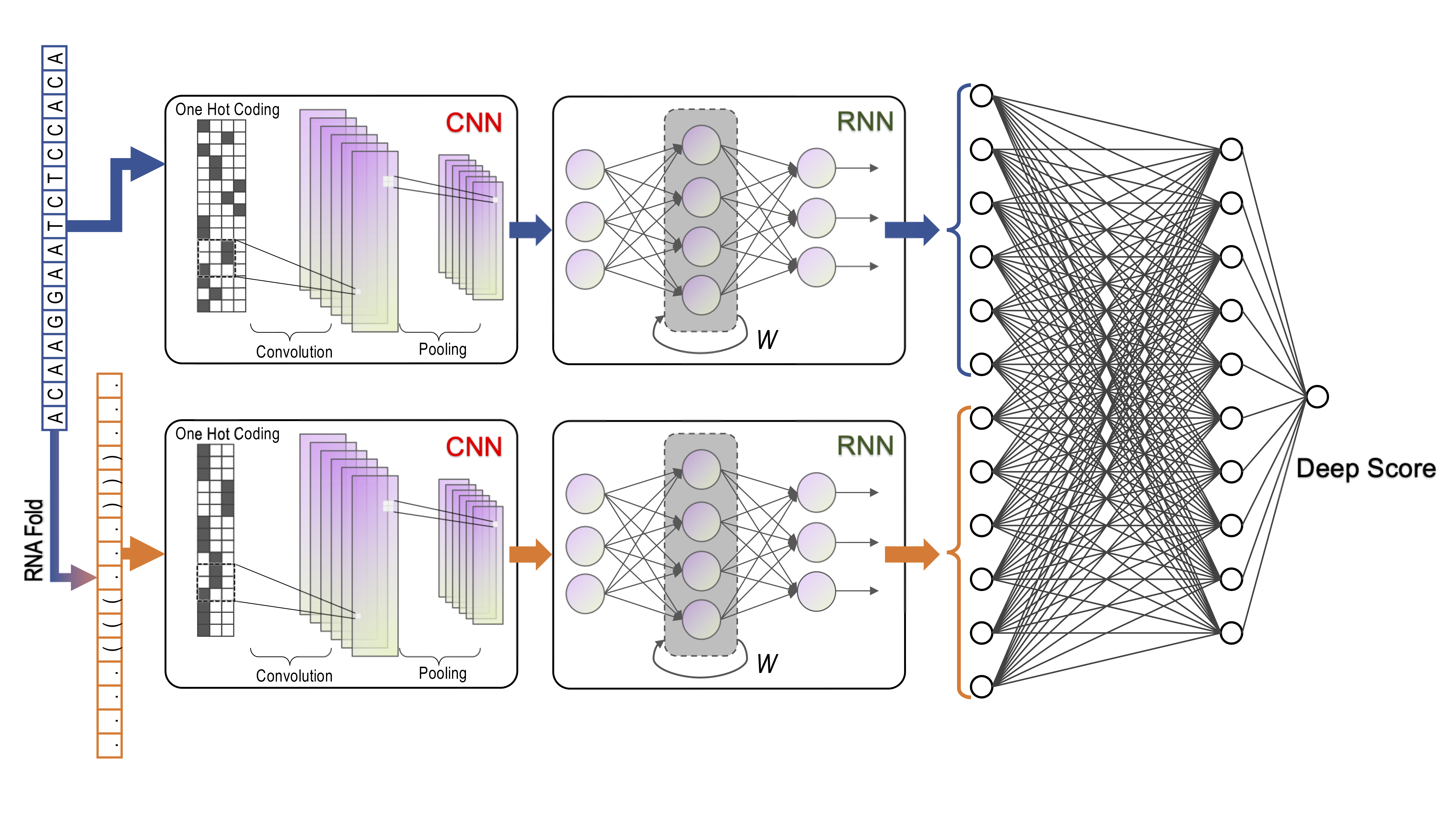
DeepCas13 is developed by Wei Li's Lab at Children's National Medical Center, which is a deep learning–based computational model for predicting the RfxCas13d (CasRx) sgRNA on-target efficiency.
DeepCas13 uses convolutional neural network (CNN) and following recurrent neural network (RNN) to extract spatial-temporal features for both sequence and secondary structure of one specific sgRNA and then predicts the efficiency by a fully connected neural network (FCNN).
What is Deep Score
Deep Score, which ranges from 0 to 1, is used to indicate the on-target efficiency of a specific sgRNA. The higher the Deep Score, the more likely sgRNA is to be effective.
What is lentiviral deficiency score
Cas13 systems under lentiviral vectors exhibit differential levels of lentiviral deficiency by which lentiviral Cas13 compoments cannot be properly packaged and delivered through lentivirus particles.
For one-vector RfxCas13d system in which Cas13d and its sgRNA cassette are in the same lentiviral vector, some crRNA inserts may elicit crRNA-specific lentiviral deficiency effect that leads to compromised lentiviral infection of Cas13d compoments into target cells.
We use the pre-trained Lasso Model to predict the potential of lentiviral deficiency for any given crRNA for RfxCas13d. The higer lentiviral deficiency score, the higher chance that the sgRNA with lentiviral deficiency effect.
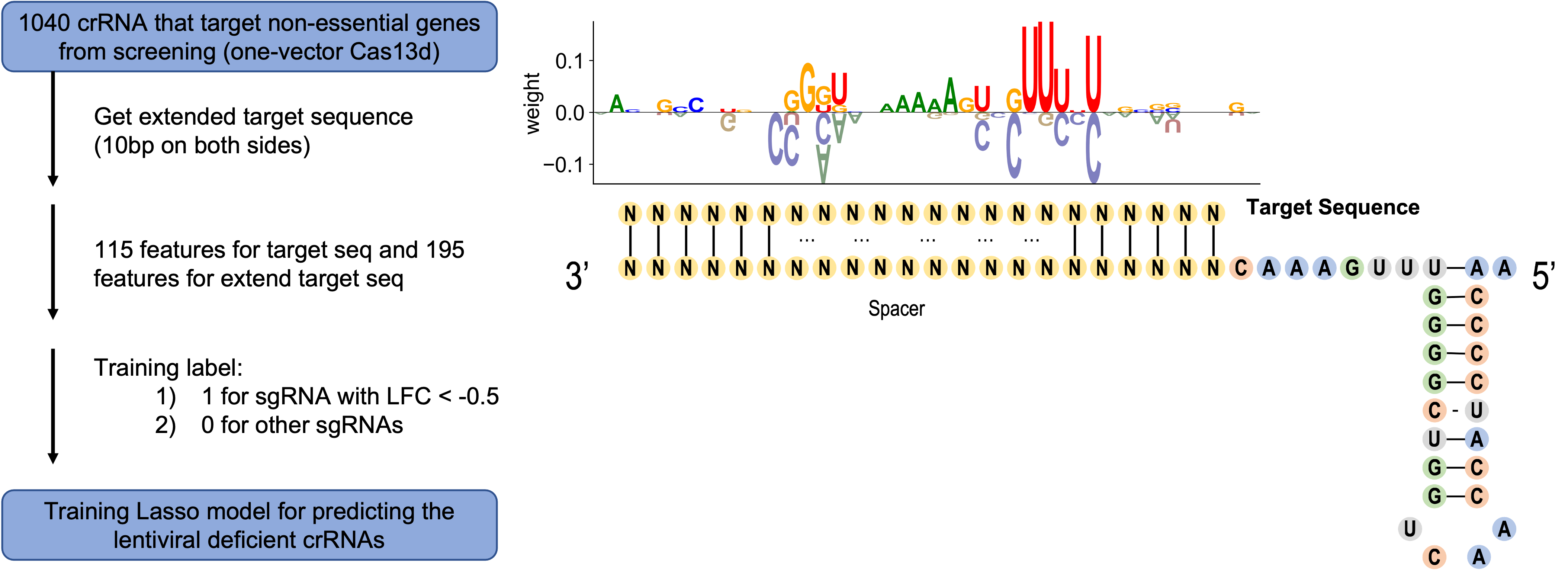
Here is how we develop the prediction model: in one-vector Cas13d screen, only the gRNAs that target non-essential genes were preserved. Among these, partial gRNAs designed for junctions were further filtered. The extended target sequence (±10 bp on both sides of the spacer region) of the remaining gRNAs were used for the lasso model training. Based on gRNA summary table of one-vector Cas13d screen (Day5 vs Plasmid), label 1 was set for those gRNAs with LFC < -0.5 and label 0 was set for other gRNAs. Totally, 195 features were generated for each extended target sequence, including one-hot coding for base type in each locus, pair bases content and melting temperature of different fragments. The coefficients (weight vector) were calculated and those base location related weights were used to show the base preference for each locus.
Contact us
For further questions or to report problems, please contact us:
• Wei Li: wli2@childrensnational.org
• Xiaolong Cheng: xcheng@childrensnational.org
• Ruocheng Shan: shanruocheng@gwu.edu